We continually collaborate with our clients to ensure their analysis goals are being met.
Whether it's standard analysis pipelines, or custom data mining or visualizations, we have the expertise to deliver reproducible results, timely.

We continually collaborate with our clients to ensure their analysis goals are being met.
Whether it's standard analysis pipelines, or custom data mining or visualizations, we have the expertise to deliver reproducible results, timely.

Treeworks is our new algorithm for manageable network construction and illuminating crosstalk between known signaling pathways.
The problem: often, within high throughput experimental data, known signaling pathways and protein-protein integration maps are incomplete. Not all of the components of each signaling cascade are identified, and thus, researchers miss both direct and indirect interactions. These missing links are often critical for the interpretation of functional significance within the data.

With Treeworks, we can connect seemingly unconnected data to find relevant interactions. For example; connecting phosphoproteomic with transcriptomic data without the seemingly linking proteomics data.
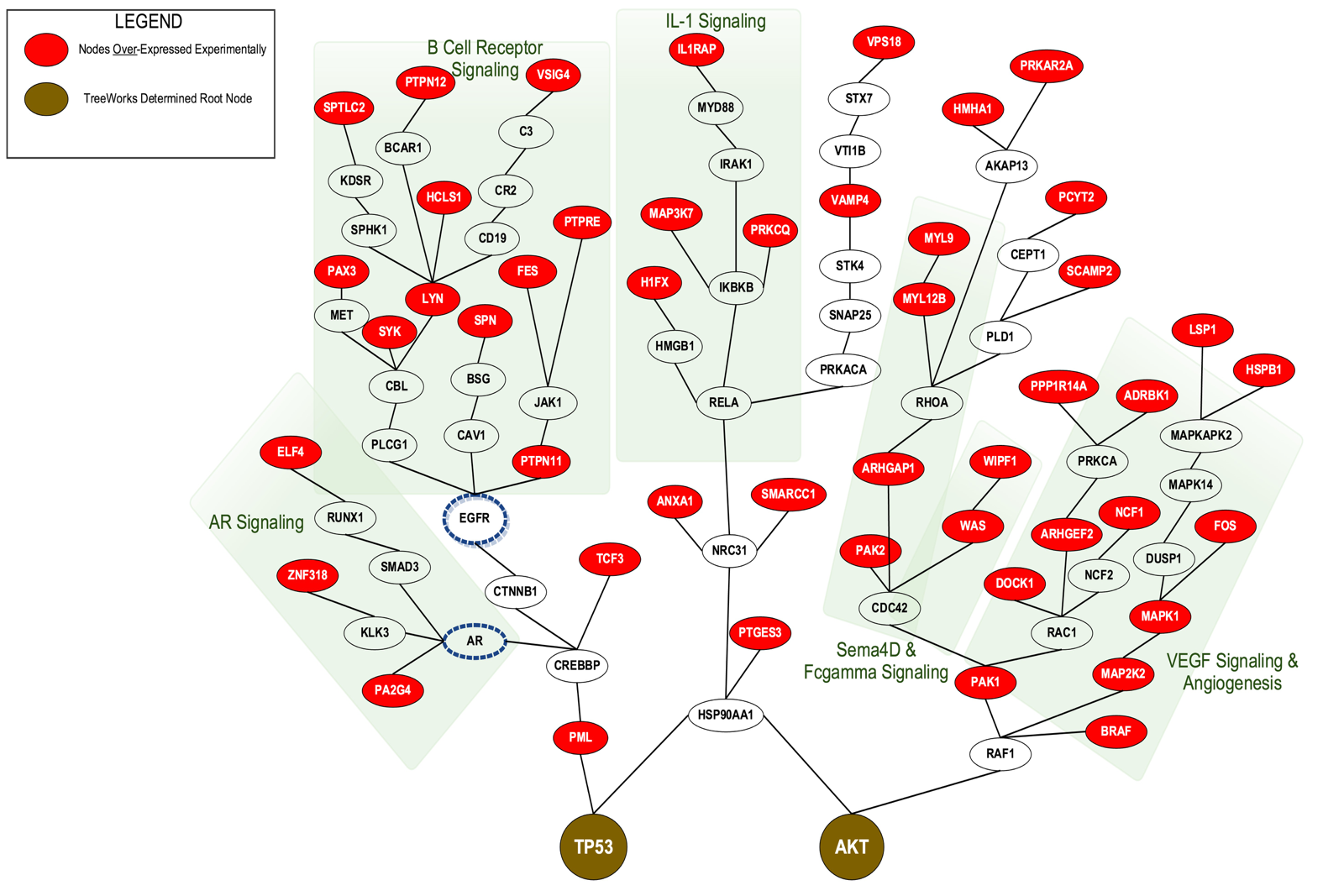
TreeWorks solution to 425 differentially expressed phosphoproteins in patient AML blasts compared with healthy control
Treeworks offers the connection between these indirect and allegedly unconnected datasets by;
Subscribe to receive a free datasheet showcasing our Treeworks methodology